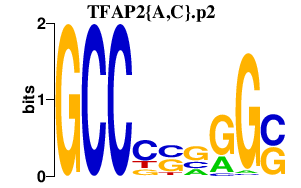
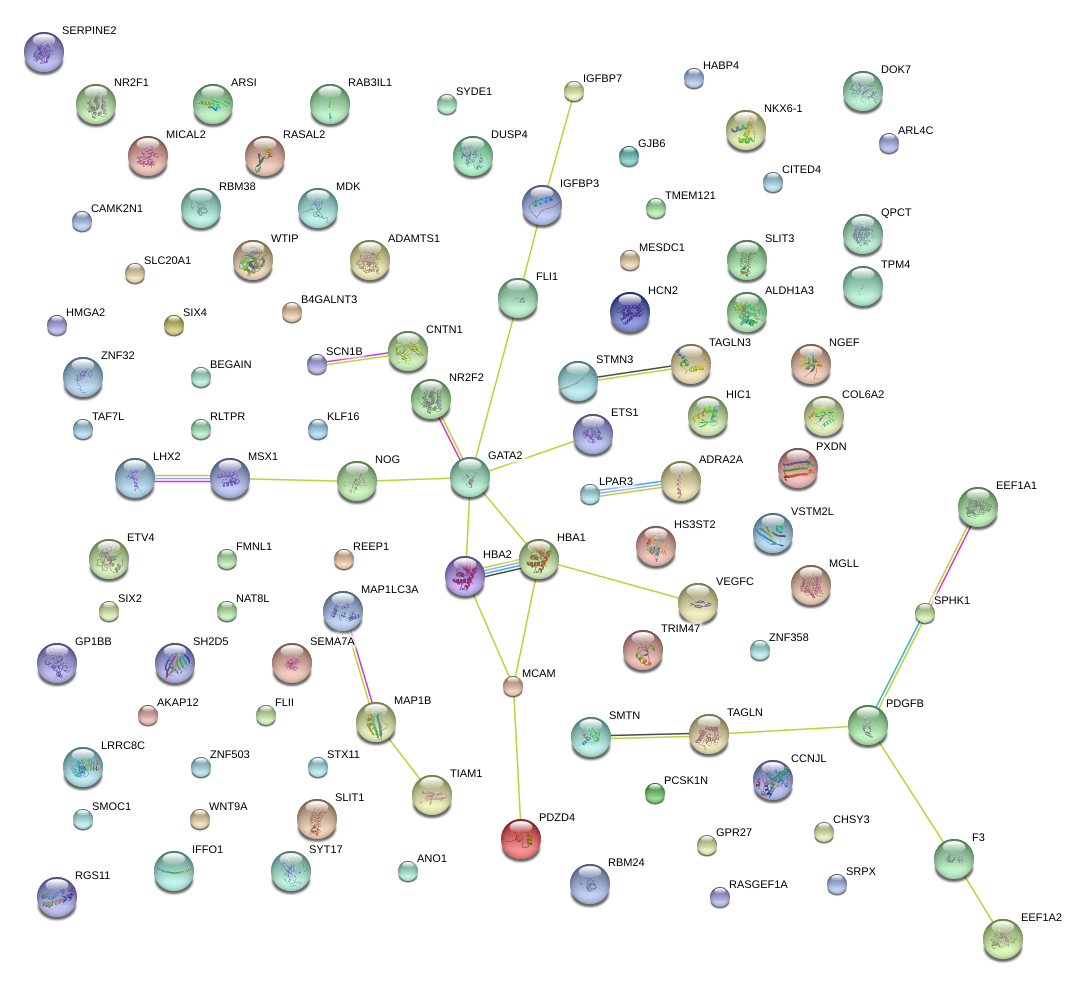
|
chr6_+_151561653
|
14.350
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr19_+_7580987
|
12.765
|
NM_018083
|
ZNF358
|
zinc finger protein 358
|
|
chr6_+_151561508
|
11.590
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr6_+_151561571
|
10.716
|
|
|
|
|
chr4_-_57976516
|
10.399
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr17_+_54671059
|
10.066
|
NM_005450
|
NOG
|
noggin
|
|
chr3_-_45267068
|
9.564
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr3_-_45267620
|
9.262
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr16_+_226678
|
8.968
|
NM_000558
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr20_+_33146500
|
8.763
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr16_+_222845
|
8.273
|
NM_000517
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr10_-_43762366
|
8.103
|
NM_145313
|
RASGEF1A
|
RasGEF domain family, member 1A
|
|
chr14_+_105992930
|
8.097
|
NM_025268
|
TMEM121
|
transmembrane protein 121
|
|
chr4_-_57976355
|
8.085
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr3_+_71803200
|
7.858
|
NM_018971
|
GPR27
|
G protein-coupled receptor 27
|
|
chr20_+_36531548
|
7.615
|
|
VSTM2L
|
V-set and transmembrane domain containing 2 like
|
|
chr16_+_22825806
|
7.265
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr5_+_92920592
|
7.198
|
|
NR2F1
NR2F2
|
nuclear receptor subfamily 2, group F, member 1
nuclear receptor subfamily 2, group F, member 2
|
|
chr20_-_62130390
|
6.835
|
NM_001958
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2
|
|
chr20_+_36531498
|
6.756
|
NM_080607
|
VSTM2L
|
V-set and transmembrane domain containing 2 like
|
|
chr11_+_69924407
|
6.666
|
NM_018043
|
ANO1
|
anoctamin 1, calcium activated chloride channel
|
|
chr11_-_128392061
|
6.617
|
NM_001162422
NM_005238
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr7_-_106301329
|
6.613
|
NM_175884
|
C7orf74
|
chromosome 7 open reading frame 74
|
|
chr5_-_168727715
|
6.556
|
NM_003062
|
SLIT3
|
slit homolog 3 (Drosophila)
|
|
chr15_+_81293286
|
6.492
|
NM_022566
|
MESDC1
|
mesoderm development candidate 1
|
|
chr11_+_12132063
|
6.444
|
NM_014632
|
MICAL2
|
microtubule associated monoxygenase, calponin and LIM domain containing 2
|
|
chr11_+_117070039
|
6.421
|
NM_001001522
NM_003186
|
TAGLN
|
transgelin
|
|
chr11_-_119187811
|
6.339
|
NM_006500
|
MCAM
|
melanoma cell adhesion molecule
|
|
chr1_-_85358851
|
6.266
|
NM_012152
|
LPAR3
|
lysophosphatidic acid receptor 3
|
|
chr5_+_129240453
|
6.246
|
NM_175856
|
CHSY3
|
chondroitin sulfate synthase 3
|
|
chr12_+_569542
|
6.186
|
NM_173593
|
B4GALNT3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3
|
|
chr1_+_90098693
|
6.176
|
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr17_+_43299277
|
6.168
|
NM_005892
|
FMNL1
|
formin-like 1
|
|
chr11_-_128391888
|
6.154
|
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr2_+_113403433
|
6.126
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr20_-_62284701
|
6.120
|
NM_015894
|
STMN3
|
stathmin-like 3
|
|
chr1_-_228135587
|
5.989
|
NM_003395
|
WNT9A
|
wingless-type MMTV integration site family, member 9A
|
|
chr16_+_67686813
|
5.952
|
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr21_-_28217272
|
5.925
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr1_-_95007139
|
5.912
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr16_+_19179534
|
5.882
|
NM_016524
|
SYT17
|
synaptotagmin XVII
|
|
chrX_-_48693934
|
5.869
|
NM_013271
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor
|
|
chrX_-_153095810
|
5.868
|
|
PDZD4
|
PDZ domain containing 4
|
|
chr8_-_29207795
|
5.851
|
NM_001394
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr9_+_99212379
|
5.769
|
NM_014282
|
HABP4
|
hyaluronan binding protein 4
|
|
chr16_-_325867
|
5.755
|
NM_003834
NM_183337
|
RGS11
|
regulator of G-protein signaling 11
|
|
chr9_+_126773851
|
5.677
|
NM_004789
|
LHX2
|
LIM homeobox 2
|
|
chr13_-_20806533
|
5.630
|
NM_001110219
NM_001110220
NM_001110221
|
GJB6
|
gap junction protein, beta 6, 30kDa
|
|
chr3_-_127541160
|
5.627
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr15_+_101420032
|
5.626
|
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr17_+_74380676
|
5.624
|
NM_001142601
NM_021972
NM_182965
|
SPHK1
|
sphingosine kinase 1
|
|
chr15_-_74726258
|
5.585
|
NM_001146029
NM_003612
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group)
|
|
chrX_-_38079980
|
5.569
|
NM_001170750
NM_001170751
NM_001170752
NM_006307
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chr12_+_66218879
|
5.538
|
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr19_+_35521591
|
5.512
|
NM_001037
NM_199037
|
SCN1B
|
sodium channel, voltage-gated, type I, beta
|
|
chr22_+_19711065
|
5.467
|
NM_000407
|
GP1BB
|
glycoprotein Ib (platelet), beta polypeptide
|
|
chr21_-_28217692
|
5.441
|
NM_006988
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr5_+_71403040
|
5.405
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr12_-_6665220
|
5.372
|
NM_001039670
NM_001193457
NM_080730
|
IFFO1
|
intermediate filament family orphan 1
|
|
chr2_-_224903933
|
5.362
|
NM_001136528
NM_006216
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr3_-_127541975
|
5.351
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr15_+_101420004
|
5.346
|
NM_000693
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr17_+_1959512
|
5.346
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr4_-_57976550
|
5.314
|
NM_001253835
NM_001553
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr4_-_177713664
|
5.295
|
NM_005429
|
VEGFC
|
vascular endothelial growth factor C
|
|
chr11_+_128564130
|
5.293
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr19_+_15218141
|
5.192
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr1_+_90098624
|
5.174
|
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr1_-_21059132
|
5.141
|
NM_001103160
NM_001103161
|
SH2D5
|
SH2 domain containing 5
|
|
chr1_-_41328002
|
5.120
|
NM_133467
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4
|
|
chr11_-_61684981
|
5.065
|
NM_013401
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1
|
|
chr7_-_45960738
|
5.062
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr14_-_101034261
|
5.039
|
NM_001159531
|
BEGAIN
|
brain-enriched guanylate kinase-associated homolog (rat)
|
|
chr21_-_32931289
|
5.032
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr4_+_2061238
|
4.999
|
NM_178557
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative)
|
|
chr1_+_90098643
|
4.977
|
NM_032270
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr14_-_61190458
|
4.974
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr10_-_44144266
|
4.963
|
NM_006973
|
ZNF32
|
zinc finger protein 32
|
|
chr19_+_16187306
|
4.956
|
|
TPM4
|
tropomyosin 4
|
|
chr19_+_16187047
|
4.876
|
NM_003290
|
TPM4
|
tropomyosin 4
|
|
chr20_+_55966440
|
4.870
|
NM_017495
NM_183425
|
RBM38
|
RNA binding motif protein 38
|
|
chr5_-_159739482
|
4.856
|
NM_024565
|
CCNJL
|
cyclin J-like
|
|
chrX_-_153095996
|
4.844
|
NM_032512
|
PDZD4
|
PDZ domain containing 4
|
|
chr22_-_39639967
|
4.836
|
|
PDGFB
|
platelet-derived growth factor beta polypeptide
|
|
chr19_+_34972866
|
4.799
|
NM_001080436
|
WTIP
|
Wilms tumor 1 interacting protein
|
|
chr6_+_144471631
|
4.797
|
NM_003764
|
STX11
|
syntaxin 11
|
|
chr19_+_16187266
|
4.786
|
|
TPM4
|
tropomyosin 4
|
|
chr3_-_128206704
|
4.769
|
NM_001145662
|
GATA2
|
GATA binding protein 2
|
|
chr12_-_93965521
|
4.750
|
|
LOC144481
|
uncharacterized LOC144481
|
|
chr3_+_111718006
|
4.748
|
NM_001008273
|
TAGLN3
|
transgelin 3
|
|
chr2_-_235405692
|
4.696
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr2_-_45236521
|
4.695
|
NM_016932
|
SIX2
|
SIX homeobox 2
|
|
chr4_+_4861371
|
4.687
|
NM_002448
|
MSX1
|
msh homeobox 1
|
|
chr4_-_85419386
|
4.670
|
NM_006168
|
NKX6-1
|
NK6 homeobox 1
|
|
chr17_-_73874634
|
4.624
|
NM_033452
|
TRIM47
|
tripartite motif containing 47
|
|
chr1_-_41328019
|
4.550
|
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4
|
|
chr1_+_178063237
|
4.528
|
|
RASAL2
|
RAS protein activator like 2
|
|
chr8_-_13133956
|
4.527
|
|
|
|
|
chr1_+_178063302
|
4.527
|
|
RASAL2
|
RAS protein activator like 2
|
|
chr10_-_77161136
|
4.525
|
|
ZNF503
|
zinc finger protein 503
|
|
chr4_+_3465032
|
4.513
|
NM_001164673
NM_173660
|
DOK7
|
docking protein 7
|
|
chr5_-_149681935
|
4.494
|
|
ARSI
|
arylsulfatase family, member I
|
|
chr14_+_70346113
|
4.451
|
NM_001034852
NM_022137
|
SMOC1
|
SPARC related modular calcium binding 1
|
|
chrX_-_153095746
|
4.448
|
|
PDZD4
|
PDZ domain containing 4
|
|
chr17_-_73874056
|
4.428
|
|
TRIM47
|
tripartite motif containing 47
|
|
chr12_+_66218141
|
4.423
|
NM_003483
NM_003484
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr19_+_589849
|
4.395
|
NM_001194
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2
|
|
chr22_+_31487315
|
4.378
|
|
SMTN
|
smoothelin
|
|
chr19_-_1863425
|
4.370
|
|
KLF16
|
Kruppel-like factor 16
|
|
chr2_-_1748285
|
4.370
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr2_+_37571752
|
4.369
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr2_-_224903324
|
4.356
|
NM_001136530
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr1_-_20812271
|
4.353
|
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr21_+_47531369
|
4.349
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr11_+_46402333
|
4.337
|
NM_001012334
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr2_-_233792825
|
4.330
|
NM_001114090
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chr17_-_41623652
|
4.322
|
NM_001079675
|
ETV4
|
ets variant 4
|
|
chr2_-_86564632
|
4.288
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr12_+_104850641
|
4.258
|
NM_001173982
NM_018413
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr6_+_41606193
|
4.256
|
NM_005586
|
MDFI
|
MyoD family inhibitor
|
|
chr20_-_25038501
|
4.252
|
NM_001252675
NM_001252677
NM_032501
|
ACSS1
|
acyl-CoA synthetase short-chain family member 1
|
|
chr2_-_20212448
|
4.240
|
NM_002381
|
MATN3
|
matrilin 3
|
|
chr7_+_128470462
|
4.237
|
NM_001127487
NM_001458
|
FLNC
|
filamin C, gamma
|
|
chr2_+_103236147
|
4.211
|
NM_003048
|
SLC9A2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 2
|
|
chr11_+_128563993
|
4.209
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr12_+_104850762
|
4.198
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr18_+_9708322
|
4.191
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr13_-_41768533
|
4.181
|
NM_032138
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7
|
|
chr5_+_133450353
|
4.178
|
NM_003202
|
TCF7
|
transcription factor 7 (T-cell specific, HMG-box)
|
|
chr19_+_16187315
|
4.163
|
|
TPM4
|
tropomyosin 4
|
|
chr2_-_1748213
|
4.118
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr5_+_34656482
|
4.106
|
|
RAI14
|
retinoic acid induced 14
|
|
chr2_-_20212408
|
4.104
|
|
MATN3
|
matrilin 3
|
|
chr17_+_1958354
|
4.093
|
NM_006497
|
HIC1
|
hypermethylated in cancer 1
|
|
chr2_-_1748317
|
4.075
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr9_+_131183122
|
4.074
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr10_-_77161539
|
4.058
|
|
ZNF503
|
zinc finger protein 503
|
|
chr8_-_133492751
|
4.025
|
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr2_-_218843541
|
4.011
|
|
TNS1
|
tensin 1
|
|
chr12_+_104850784
|
4.004
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr8_-_133493003
|
4.000
|
NM_004519
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr12_+_104850750
|
3.997
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr10_-_99094264
|
3.960
|
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2
|
|
chrX_+_68725077
|
3.959
|
NM_015686
|
FAM155B
|
family with sequence similarity 155, member B
|
|
chr12_+_58005217
|
3.947
|
NM_182947
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25
|
|
chr22_-_20792107
|
3.943
|
NM_153334
NM_182895
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr12_+_53491448
|
3.937
|
|
IGFBP6
|
insulin-like growth factor binding protein 6
|
|
chr12_+_111471827
|
3.936
|
NM_015267
|
CUX2
|
cut-like homeobox 2
|
|
chr19_+_39989603
|
3.919
|
|
DLL3
|
delta-like 3 (Drosophila)
|
|
chr19_-_1863546
|
3.918
|
NM_031918
|
KLF16
|
Kruppel-like factor 16
|
|
chr10_+_17271859
|
3.917
|
|
VIM
|
vimentin
|
|
chr22_+_51113069
|
3.916
|
NM_001080420
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3
|
|
chr16_+_89989686
|
3.916
|
NM_006086
|
TUBB3
|
tubulin, beta 3 class III
|
|
chr15_+_41786055
|
3.909
|
NM_002220
|
ITPKA
|
inositol-trisphosphate 3-kinase A
|
|
chr2_+_231577624
|
3.900
|
|
CAB39
|
calcium binding protein 39
|
|
chr3_-_197282805
|
3.888
|
NM_004051
NM_203314
|
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1
|
|
chr17_-_46703613
|
3.885
|
NM_024017
|
HOXB9
|
homeobox B9
|
|
chr12_+_104850986
|
3.879
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr22_+_31489343
|
3.875
|
|
SMTN
|
smoothelin
|
|
chr1_+_23037329
|
3.859
|
NM_004442
NM_017449
|
EPHB2
|
EPH receptor B2
|
|
chr3_+_111717585
|
3.847
|
NM_001008272
NM_013259
|
TAGLN3
|
transgelin 3
|
|
chr8_+_22436641
|
3.838
|
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr8_-_38325524
|
3.824
|
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr5_+_34656367
|
3.824
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr17_+_74864427
|
3.816
|
NM_001199172
NM_144677
|
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B
|
|
chr10_+_17271956
|
3.808
|
|
VIM
|
vimentin
|
|
chr11_-_2950593
|
3.795
|
NM_003311
|
PHLDA2
|
pleckstrin homology-like domain, family A, member 2
|
|
chr2_+_231577536
|
3.790
|
NM_016289
|
CAB39
|
calcium binding protein 39
|
|
chr10_-_77161424
|
3.787
|
NM_032772
|
ZNF503
|
zinc finger protein 503
|
|
chr2_-_43453724
|
3.782
|
|
ZFP36L2
|
zinc finger protein 36, C3H type-like 2
|
|
chr17_+_64960754
|
3.779
|
NM_014405
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4
|
|
chr5_+_140346289
|
3.765
|
NM_031883
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|
|
chr1_+_92495484
|
3.758
|
NM_173567
|
EPHX4
|
epoxide hydrolase 4
|
|
chr4_+_1005395
|
3.752
|
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr15_-_65067696
|
3.748
|
NM_194272
|
RBPMS2
|
RNA binding protein with multiple splicing 2
|
|
chr12_-_71003622
|
3.744
|
NM_001206971
NM_001206972
NM_002837
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr9_-_86153048
|
3.743
|
NM_001244959
NM_174938
|
FRMD3
|
FERM domain containing 3
|
|
chr6_+_151561094
|
3.710
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr21_+_47518012
|
3.705
|
NM_001849
NM_058174
NM_058175
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr19_+_35168558
|
3.693
|
NM_001012320
NM_018443
|
ZNF302
|
zinc finger protein 302
|
|
chr19_-_41859830
|
3.648
|
NM_000660
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr5_-_121413967
|
3.640
|
|
LOX
|
lysyl oxidase
|
|
chr8_+_145582267
|
3.629
|
NM_024531
|
GPR172A
|
G protein-coupled receptor 172A
|
|
chr9_+_131644427
|
3.609
|
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr22_+_31031672
|
3.596
|
NM_001001479
|
SLC35E4
|
solute carrier family 35, member E4
|
|
chr9_+_131644368
|
3.587
|
NM_001127245
NM_019594
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr10_+_64133915
|
3.578
|
NM_014951
NM_199450
NM_199451
|
ZNF365
|
zinc finger protein 365
|
|
chr16_+_57126439
|
3.564
|
NM_152727
|
CPNE2
|
copine II
|
|
chr15_+_73976537
|
3.558
|
|
|
|
|
chr2_-_43453146
|
3.548
|
|
ZFP36L2
|
zinc finger protein 36, C3H type-like 2
|
|
chr3_-_128207366
|
3.544
|
NM_001145661
|
GATA2
|
GATA binding protein 2
|
|
chr21_+_38071990
|
3.536
|
NM_005069
NM_009586
|
SIM2
|
single-minded homolog 2 (Drosophila)
|
|
chr8_+_56792392
|
3.528
|
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr10_+_104404195
|
3.524
|
NM_030912
|
TRIM8
|
tripartite motif containing 8
|
|
chr2_+_239756672
|
3.523
|
NM_057179
|
TWIST2
|
twist homolog 2 (Drosophila)
|
|
chr16_+_57126494
|
3.511
|
|
CPNE2
|
copine II
|
|
chr2_+_173292344
|
3.507
|
|
ITGA6
|
integrin, alpha 6
|
|
chr1_+_178062955
|
3.500
|
|
RASAL2
|
RAS protein activator like 2
|
|
chr11_+_94277005
|
3.497
|
NM_002033
|
PIWIL4
FUT4
|
piwi-like 4 (Drosophila)
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific)
|
|
chrX_-_153151537
|
3.495
|
|
|
|